Cell-Resolved Histone and TF Profiling at Production Scale
Epigenome Technologies delivers single-cell CUT&Tag programs that profile histone modifications, transcription factors, and chromatin-associated proteins in heterogeneous populations without flow sorting. We qualify antibodies, optimize nuclei handling, and return interpretable cell-by-peak matrices that integrate directly with your scRNA-seq pipelines.
Targeted Chromatin Profiling
Precise protein–DNA mapping
Antibody-Driven Assay Design
Optimized for each target
Built for Cellular Resolution
Cell-state–specific insight
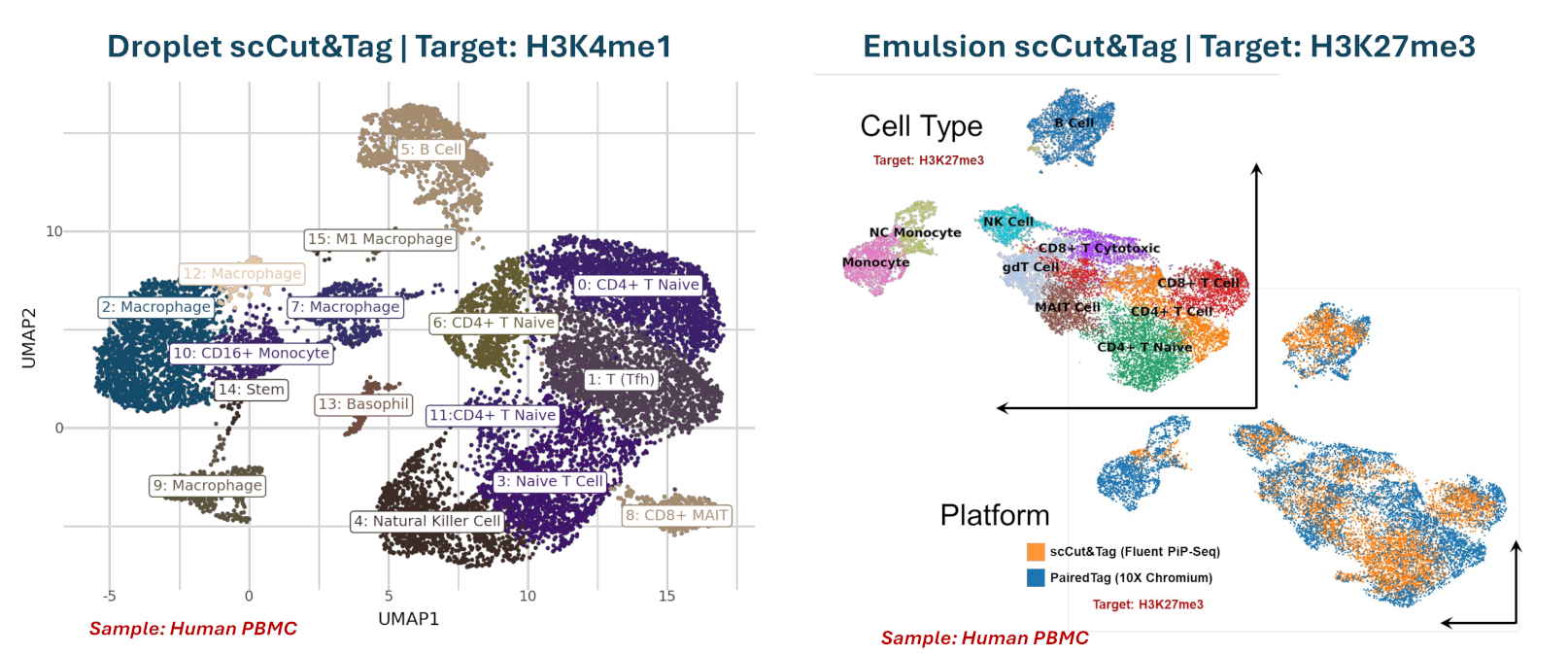
scCUT&Tag for cell-type epigenetic profiling
Single-cell CUT&Tag uses antibody-targeted tagmentation to map where histone modifications, transcription factors, or chromatin-associated proteins bind DNA across individual cells. The approach delivers fragment-level precision for regulatory element annotation, heterogeneity discovery, and rare-cell epigenetic characterization without flow cytometry pre-enrichment.
Where teams use the program
- Oncology and immunology studies requiring cell-type enhancer or silencing profiles.
- Developmental cohorts tracking dynamic histone mark transitions.
- Rare-cell epigenetic characterization in mixed tissues without FACS.
- Perturbation screens validating chromatin-state responses to CRISPR or compound treatments.
- Biomarker discovery linking epigenetic signatures to treatment response or resistance.
Collaboration model
- Joint design sessions align target antibodies, cell counts, sequencing depth, and optional RNA integration.
- Antibody validation runs confirm signal quality before committing full cohorts.
- Project scientists remain embedded through QC reviews, data interpretation, and reporting.
Included interpretation
- Cell-type clustering and epigenetic signature annotation.
- Differential peak analysis and motif enrichment summaries.
- Regulatory element linkage to scRNA-seq or bulk expression datasets.
- Actionable slide-ready visualizations delivered alongside raw outputs.
Aligning platform to experimental design
| Platform | Cat. No. | Best For | Typical Cell Count |
|---|---|---|---|
| Droplet scCUT&Tag | SCTDS201 | 10x Genomics-compatible workflows; integrated multiome readouts | 5,000–10,000 cells/sample |
| BD Rhapsody compatible scCUT&Tag | SCTBS201 | BD Rhapsody-compatible workflows; high cel recovery | 30,000–80,000 cells/sample |
| Emulsion scCUT&Tag | SCTDE201 | High-throughput cohorts; flexible cell input and antibody panels | 10,000–50,000 cells/sample |
Droplet scCUT&Tag

Integrates with 10x Genomics platforms for straightforward scRNA-seq + epigenetic multiome workflows. Ideal when combining transcript and histone modification profiles in the same cells.
- 10x-compatible barcoding for direct integration with scRNA-seq
- Streamlined library construction with validated antibody panels
- Recommended for cohorts requiring 5,000–10,000 cells per sample
Emulsion scCUT&Tag

High-throughput emulsion chemistry for large-scale profiling or when 10x compatibility is not required. Flexible antibody panels and cell counts support discovery-phase heterogeneity studies.
- Scale to 2,000–50,000 cells per sample
- Custom antibody panels for non-standard histone marks or TFs
- Optimized for cohorts emphasizing epigenetic diversity over RNA linkage
Selection guidance
- Choose Droplet scCUT&Tag when integrating with existing 10x scRNA-seq workflows or requiring RNA+epigenetic co-profiling.
- Choose Emulsion scCUT&Tag for pilot or large-scale profiling, or when RNA linkage is not the primary objective.
Transparent workflow and checkpoints
-
Plan & validate antibodies
We confirm target antibodies, cell counts, and nuclei handling protocols; optional antibody validation runs ensure signal quality before full cohort commitment.
-
Prepare & construct
Nuclei isolation, antibody staining, and droplet or emulsion encapsulation with QC on viability, tagmentation efficiency, and fragment distribution.
-
Sequence
Paired-end runs on NovaSeq or NextSeq platforms with depth monitoring against internal signal-to-noise benchmarks.
-
Interpret
Automated cell calling, peak annotation, and cell-type clustering with data packages aligned to your preferred analysis environment.
Operational cadence
- Day 3 antibody validation QC review (if requested) with your project scientist.
- Day 7 nuclei prep and library construction status with preliminary fragment profiles.
- Day 12 library acceptance with complexity and tagmentation efficiency reports.
- Day 24 FASTQ and cell-by-peak matrices delivered; interpretive briefing.
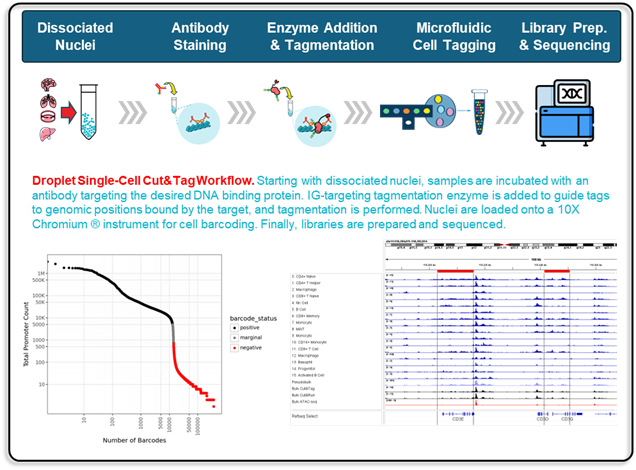
Top-Tier Quality Metrics
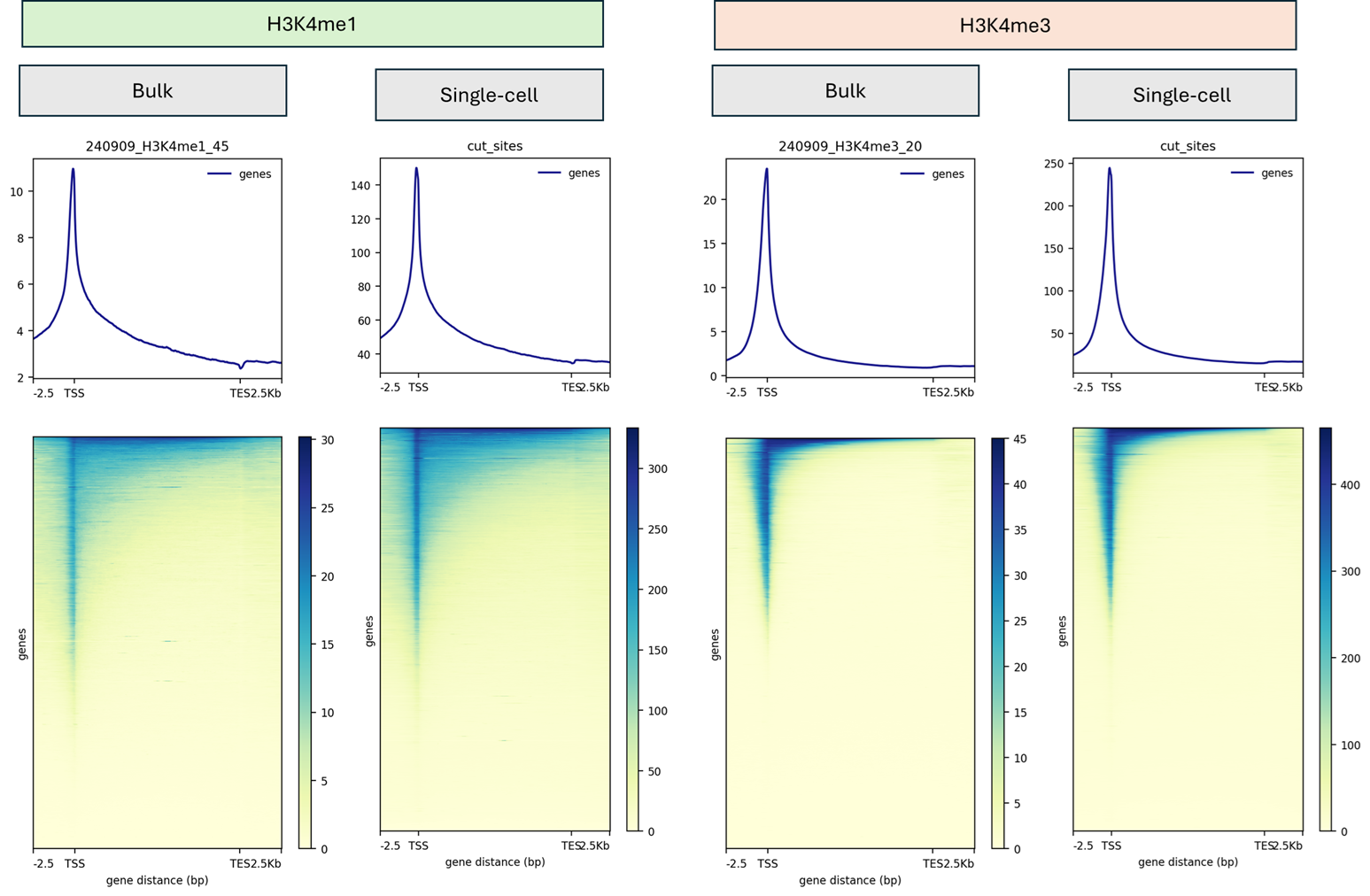
- TSS enrichment distributions with cohort medians and antibody-specific benchmarks.
- Fragment size and nucleosome phasing profiles reviewed prior to sequencing approval.
- Cell-barcode complexity and background noise assessments for every sample.
- Signal-to-noise overlays compared to internal gold standards and published scCUT&Tag references.
Delivery package
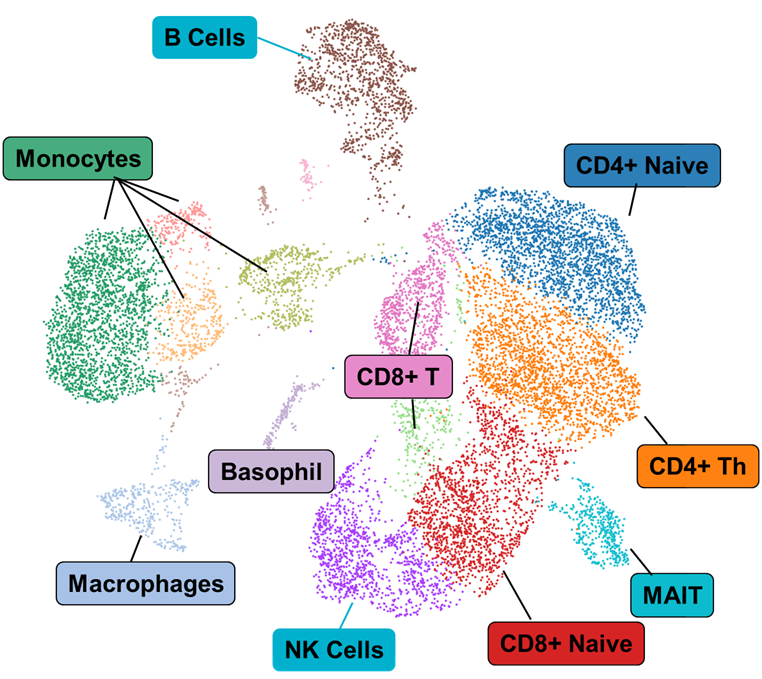
- Sequencing-ready libraries with concentration, fragment distribution, and complexity documentation.
- FASTQ, cell-by-peak matrices, and metadata formatted for Seurat, Scanpy, or custom pipelines.
- Optional interpretive report summarizing cell-type clustering, differential peaks, and regulatory element annotations.
Partner with our scientists
Share your target antibodies, cohort size, and required timelines. We will return a scoped scCUT&Tag brief outlining platform selection, antibody validation strategy, QC checkpoints, and downstream reporting.
- Sample requirements: 100K–200K nuclei preferred depending on platform; low-input contingencies available.
- Storage guidance: Fresh or cryopreserved nuclei accepted with documented handling.
- Data options: Raw, processed, and interpretive outputs available individually or bundled.
- Support: Project scientists provide experimental planning and guidance, data reviews, and troubleshooting.