scCUT&Tag Kits — Single-Cell Epigenetic Profiling
Profile histone modifications, transcription factors, and chromatin-associated proteins in individual cells without flow sorting. Flexible antibody targeting across 10x Chromium, BD Rhapsody, and Illumina PIPseq platforms. Ultra-low input, production-ready quality control, and expert support included.
Cell Recovery
70–90%
Quality Gate
Optimized QC
Platforms
3 Options
Choose Your Kit

| Product | Catalogue # | Platform | Size | Action |
|---|---|---|---|---|
| Droplet scCUT&Tag | SCT8101 | 10x Chromium | 8 Reactions | Order |
| BD Rhapsody-compatible scCUT&Tag | BCT8101 | BD Rhapsody | 8 Reactions | Order |
| Emulsion scCUT&Tag | ECT4101 | Illumina PIPseq | 4 Reactions | Order |
| scCUT&Tag2K | SC2K8101 | Illumina PIPseq 2K | 8 Reactions | Pre-Order |
Need guidance? Compare all kit options →
What You Need to Get Started
- Validated Antibody: Primary antibody targeting your histone modification, transcription factor, or chromatin protein of interest
- Single-Cell Platform: 10x Chromium ATAC system, BD Rhapsody ATAC, or Illumina PIPseq (choose your platform)
- Reagents: Platform-appropriate library prep kit (10x ATAC, BD ATAC, or PIPseq)
- Sequencing: Illumina-compatible sequencer (NovaSeq, NextSeq, or MiSeq supported)
- Analysis Pipeline: Preferred analysis environment (Python/Scanpy, R/Seurat, ArchR, or custom)
scCUT&Tag: Single-Cell Epigenetic Profiling
Single-Cell CUT&Tag is an innovative assay that precisely measures epigenetic modifications in individual cells by targeting histone post-translational modifications (PTMs) or transcription factors (TFs). Unlike multiomic approaches that capture RNA, scCUT&Tag focuses solely on tagmented DNA, allowing for deeper coverage of epigenetic marks without the complexity of concurrent transcriptomic profiling.
Core Strengths
- Platform compatibility: Seamlessly integrate with 10x Chromium, BD Rhapsody, or Illumina PIPseq systems.
- Flexible targeting: Work with any validated antibody for histone modifications, transcription factors, or chromatin remodelers.
- High sensitivity: Single-cell resolution without flow cytometry pre-enrichment or sorting.
- Sample flexibility: Fresh, frozen, cryopreserved, or even archival FFPE samples supported.
- Streamlined workflows: Pre-optimized protocols for leading single-cell systems reduce setup burden.

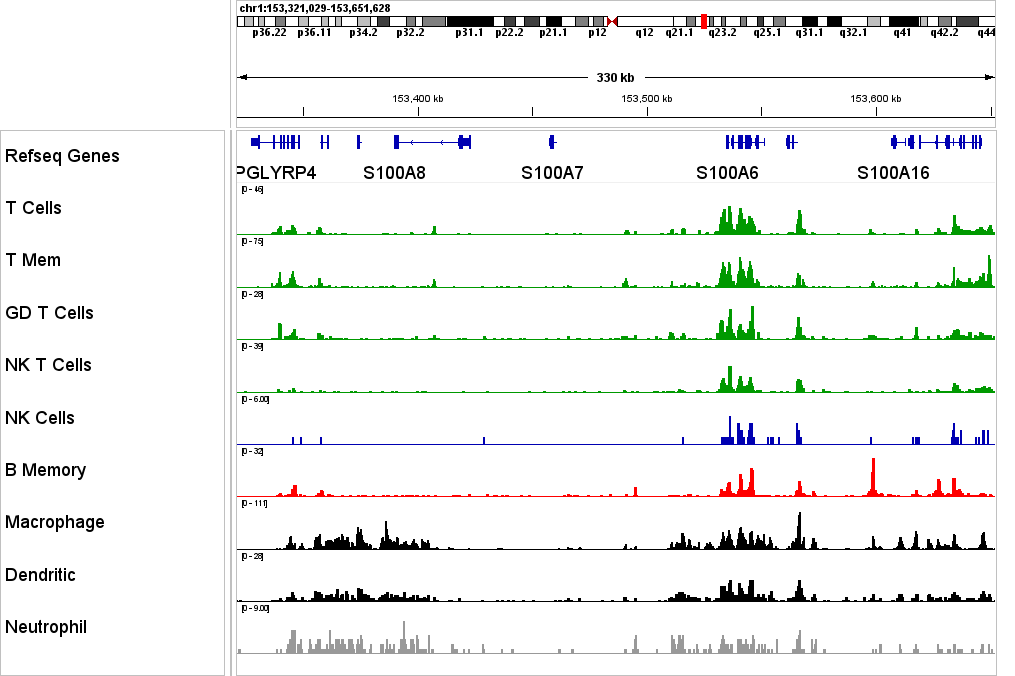
Ideal For
- Cell differentiation assays and developmental biology studies tracking epigenetic transitions.
- Drug-treatment experiments and perturbation screens measuring chromatin-state responses.
- Studies using archival FFPE tissue or other degraded RNA samples.
- Discovery of cell-type–specific regulatory programs in complex, heterogeneous tissues.
- Biomarker discovery linking epigenetic signatures to disease or treatment response.
Research Applications
Researchers use scCUT&Tag across developmental biology, immunology, cancer genomics, and perturbation studies to map epigenetic heterogeneity in complex tissues without flow cytometry.
Developmental Biology
Map epigenetic transitions across developmental stages and lineages. Identify cell-type–specific enhancer activity and histone mark signatures driving cell fate decisions.
Immunology & Cancer
Profile chromatin state heterogeneity in immune populations and tumor microenvironments. Link epigenetic signatures to treatment response and regulatory function.
Perturbation Studies
Validate chromatin-state responses to CRISPR knockout, drug treatment, or differentiation stimuli. Single-cell resolution without sorting or enrichment bias.

Choosing the Right Platform
All three platforms deliver high-quality scCUT&Tag data. Use this guide to select the platform best suited to your experimental goals, budget, and available instrumentation.
| Criteria | 10x Chromium | BD Rhapsody | Illumina PIPseq |
|---|---|---|---|
| Best For | Established 10x workflows, RNA+epigenetic multiomics | High-throughput discovery, large populations | Platform-free flexibility, budget-conscious projects |
| Cell Input Range | 5,000–10,000 typical | 10,000–50,000+ possible | Scalable 2,000–100,000 (choose kit size) |
| Median UMI/Cell | 7,000–15,000 | 8,000–35,000 | 5,000–13,000 |
| Cost per Reaction | ~$310 | ~$280 (scales favorably) | ~$150–250 (lowest) |
| Equipment Required | 10x Chromium system | BD Rhapsody system | Standard thermocycler + liquid handler |
| Data Quality | Excellent; 10x-optimized | Excellent; large-scale validated | Excellent; cost-comparable |
Choose 10x Chromium If You:
- Have existing 10x infrastructure
- Need RNA+epigenetic integration (Multiome)
- Want established, widely-validated workflows
- Require rapid turnaround (3–4 weeks)
Choose BD Rhapsody If You:
- Need maximum cell throughput (10,000–50,000+)
- Prioritize population-scale discovery
- Have BD system infrastructure
- Want cost efficiency at large scale
Choose Illumina PIPSeq If You:
- Lack dedicated droplet instrumentation
- Prioritize cost per reaction
- Want maximum experimental flexibility
- Are exploring methods or running pilots
Safety Data Sheets (SDS)
Droplet scCUT&Tag + Emulsion scCUT&Tag
- CR1007 Epigenome Technologies Quench Buffer
- CR1001 Epigenome Technologies Incubation Buffer
- CR1006 Epigenome Technologies Activator
- CF1003 Epigenome Technologies Permeabilizer
- CF1005 Epigenome Technologies Stabilizer Buffer
- CF1004 Epigenome Technologies RNAse Inhibitor
- CF1008 Epigenome Technologies Prepared TAG Enzyme
Emulsion scCUT&Tag Only
- CF1011 Epigenome Technologies Emulsion Polymerase
- CF1012 Epigenome Technologies Emulsion Ligase
- CF1013 Epigenome Technologies Emulsion ATP
- CF1014 Epigenome Technologies Master Mix
- CF1015 Epigenome Technologies Emulsion Pre-Amp Primers
- CF1016 Epigenome Technologies Emulsion Library Primers Set A
- CF1017 Epigenome Technologies Emulsion Library Primers Set B
Request Protocols & Documentation
Access detailed protocols and technical documentation by registering with us:
Product Details
Droplet scCUT&Tag — 10x Chromium Compatible
Droplet-based scCUT&Tag enables high-throughput, single-cell profiling of epigenetic marks using the familiar 10x Chromium ATAC system. Ideal for mid-to-large scale projects requiring scalable, cell-resolved views of chromatin regulation with established workflows.
Specifications
- Platform: 10x Genomics Chromium
- Typical cell yield: 5,000–10,000 cells/sample
- Median UMI/cell: 7,000–15,000 for abundant marks
- Turnaround: 3–4 weeks (kit timeline)
- Cost: ~$310 per reaction
Performance Highlights
- High-complexity libraries rival or exceed scATAC-seq for abundant marks (H3K4me1, H3K27ac).
- Optimized tagmentation chemistry preserves fragment capture while keeping background low.
- Barcode-per-nucleus strategy maintains replicate-to-replicate reproducibility.
- Delivers quantitative regulatory readouts suitable for enhancer or promoter-centric analyses.


Advantages
- High library complexity and robust per-cell quantification.
- Seamless integration with 10x ATAC and Multiome workflows.
- Clean regulatory clustering and distinct cell-type separation.
- Compatible with standard analysis tools (Scanpy, Seurat, ArchR).
- Multiomic-ready for paired RNA profiling.
Biological Applications
- Resolve lineage-specific enhancer programs and transcription-factor occupancy.
- Integrate chromatin data with scRNA-seq or scATAC-seq for multiomic interpretation.
- Track chromatin responses to perturbations, drug treatments, or differentiation cues.


Kickstart Your Droplet Project
Combine single-cell precision with 10x workflow familiarity. Our team supports antibody validation, nuclei prep, sequencing, and analysis-ready deliverables.
BD Rhapsody scCUT&Tag — High-Throughput Profiling
BD Rhapsody–based scCUT&Tag extends single-cell chromatin profiling to tens of thousands of individual cells using micro-well barcoding. Ideal for population-scale studies requiring comprehensive sampling of cellular heterogeneity across complex tissues.

Specifications
- Platform: BD Rhapsody ATAC
- Typical cell yield: 10,000–50,000+ cells/sample
- Median UMI/cell: 8,000–35,000 (platform-dependent)
- Turnaround: 4–6 weeks (kit timeline)
- Cost: ~$280 per reaction (cost-efficient at scale)
Advantages
- Massively parallel profiling of tens of thousands of nuclei.
- Efficient library construction with high UMI recovery.
- Flexible input compatibility (fresh, frozen, or sorted).
- Ideal for large discovery screens and population mapping.
- Cost-effective per-cell resolution at scale.
Why BD Rhapsody Scales
- Micro-well barcoding captures tens of thousands of nuclei per run.
- BD ATAC chemistry maintains high signal-to-noise even at large scale.
- Supports primary cells, cryopreserved material, and sorted populations.
- Ideal for population mapping and comprehensive discovery studies.
Applications
- Interrogate heterogeneous tissues (immune, neural, developmental) at scale.
- Run perturbation or compound screens that demand broad sampling.
- Profile patient cohorts where RNA quality is variable but chromatin is intact.

Best For
Large tissue samples, perturbation screens, comprehensive lineage mapping, or studies prioritizing cell number and population breadth over individual cell depth.


Launch a BD Rhapsody Project
Partner with us to translate BD Rhapsody throughput into actionable chromatin maps. We handle nuclei prep, platform setup, sequencing, and integrative analysis.
Illumina PIPseq scCUT&Tag — Platform-Free Flexibility
Illumina PIPseq–based scCUT&Tag provides flexible, platform-independent single-cell profiling using emulsion-based barcoding. Scalable from a few thousand to 100,000+ nuclei without dependence on proprietary systems, making it ideal for exploratory studies or labs without dedicated droplet instrumentation.

Specifications
- Platform: Illumina PIPseq (T2, T20, or T100)
- Typical cell yield: Flexible: 2,000–100,000 nuclei
- Median UMI/cell: 5,000–13,000 (variable by kit)
- Turnaround: 2–4 weeks (kit timeline)
- Cost: ~$150–250 per reaction (lowest per-cell cost)
Flexible Profiling Modes
- Combine chromatin with surface proteins via antibody tagging workflows.
- Plate-based isolation protects fragile or rare cell types through preprocessing.
- Swap antibody panels between runs to rapidly iterate hypotheses.
Advantages
- Platform-free: No 10x or BD system required; runs on standard equipment.
- Flexible scalability: Choose kit size matching your experiment scope.
- Cost-effective: Lowest per-reaction and per-cell cost among all platforms.
- Method flexibility: Enables rapid iteration and custom adaptations.
- Accessible to all labs: No platform lock-in or system investment.
Applications
- Flow cytometry-informed discovery of rare immune subsets.
- Plate-based CRISPR screens demanding direct chromatin readout.
- Biomarker validation pairing protein expression with enhancer state.
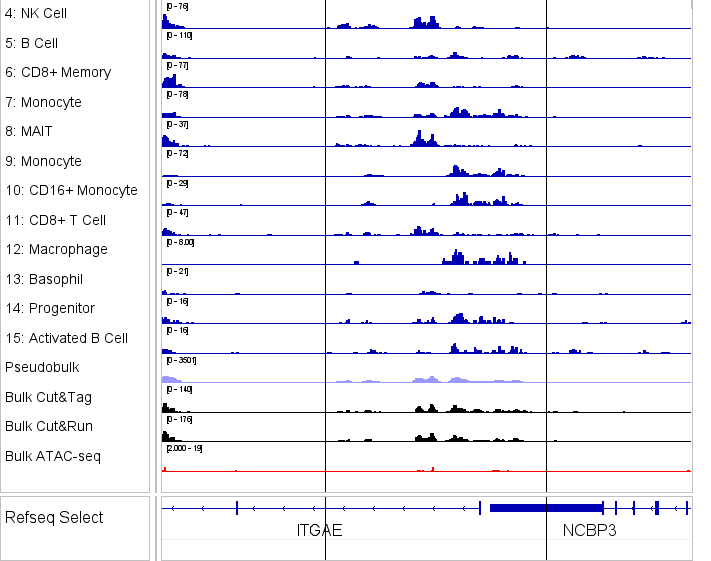

Best For
Exploratory studies, method development, budget-conscious projects, labs without dedicated droplet systems, or researchers wanting maximum flexibility without platform constraints.
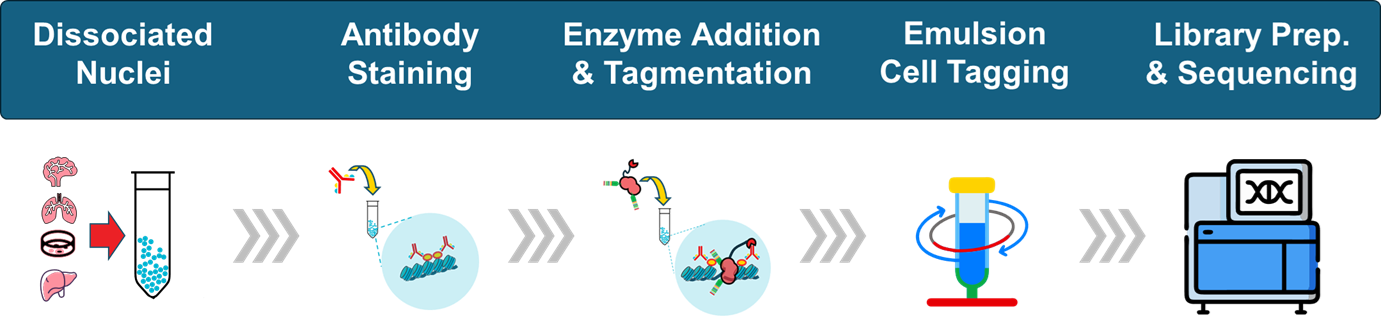
Plan Your PIPseq Study
We provide antibody panel optimization, plate layout design, and integrated sequencing analysis to accelerate PIPseq projects from pilot through scale-up.
Ready to Profile Single-Cell Epigenomics?
Start with a kit for immediate exploration, or let our experts handle the complete workflow as a managed service.