Explore Our Latest Research
Discover groundbreaking advancements in single-cell gene regulation at Epigenome Technologies. Dive into our collection of conference posters showcasing cutting-edge discoveries in biomarker identification, immune cell heterogeneity, and more.

Single-Cell Epigenetic Platforms Reveal Cell-Specific Chromatin States and Regulatory Networks in Adaptive Immunity
Using generated profiles RNA and four histone marks in over 70,000 human PBMCs, we identified thousands of interactions between chromatin state modifications and RNA expression. We demonstrated that Paired-Tag data can be used to annotate single-cell CUT&Tag data (without RNA) and demonstrated that profiles were consistent across single-cell platforms. We introduced chromatin state footprinting to assess genome-wide transcription factor states. Application to IRF binding sites revealed distinct regulatory roles across immune subsets, including naïve elements likely involved in early immune responses.
Download Poster (PDF)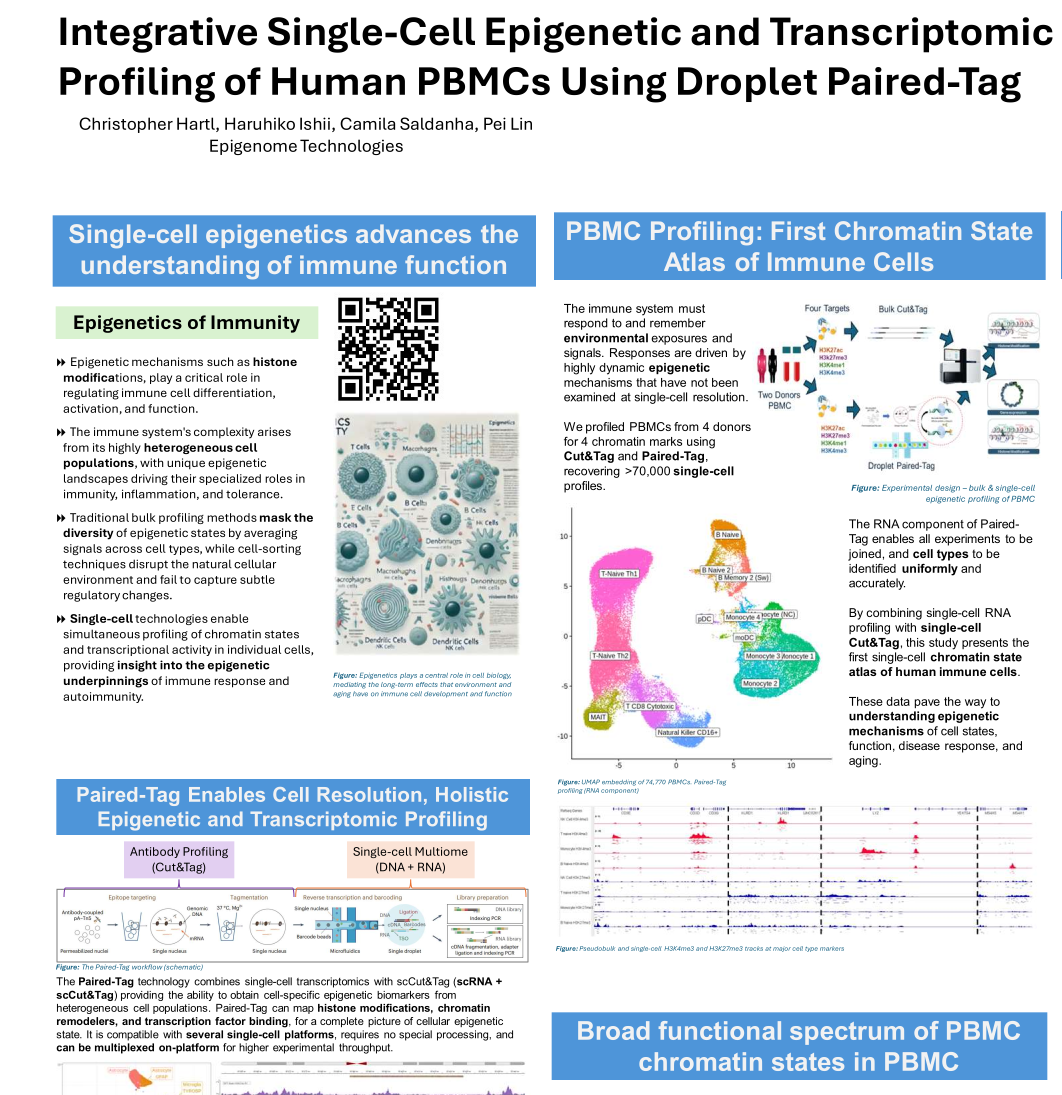
Integrative Single-Cell Epigenetic and Transcriptomic Profiling of Human PBMCs Using Droplet Paired-Tag
We profiled PBMCs from two donors using Cut&Tag and Droplet Paired-Tag, generating over 70,000 single-cell profiles. We identified hundreds of thousands of cis-regulatory elements (cCREs), primarily repressive, active, or bivalent. Most cCREs were shared across immune cell types, with a few hundred defining subtypes. Single-cell data confirmed bivalency at specific loci and showed strong correlation with bulk Cut&Tag. These epigenetic landscapes in brain and PBMCs enhance our understanding of immune cell regulation and heterogeneity.
Download Poster (PDF)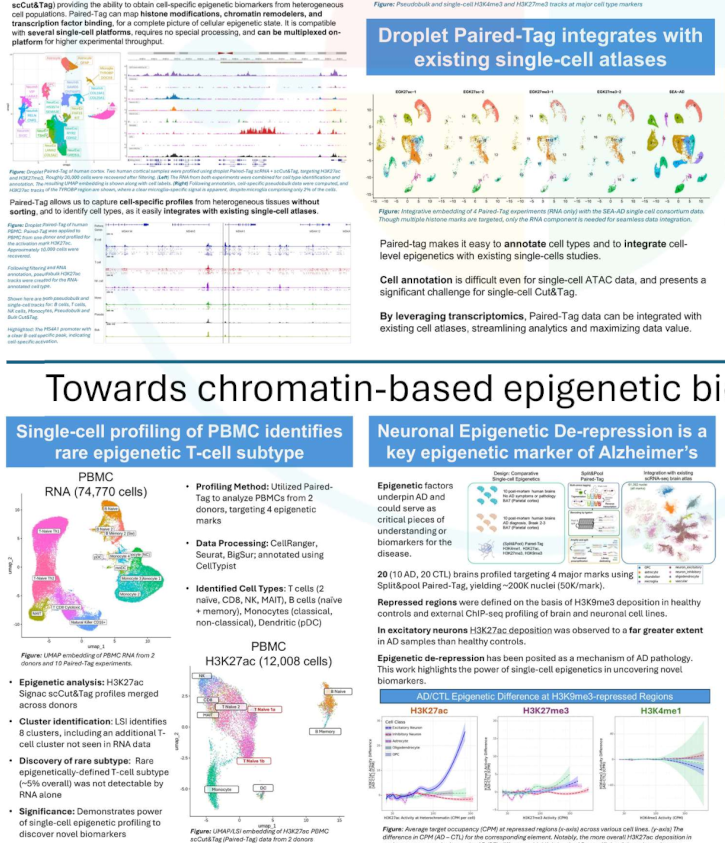
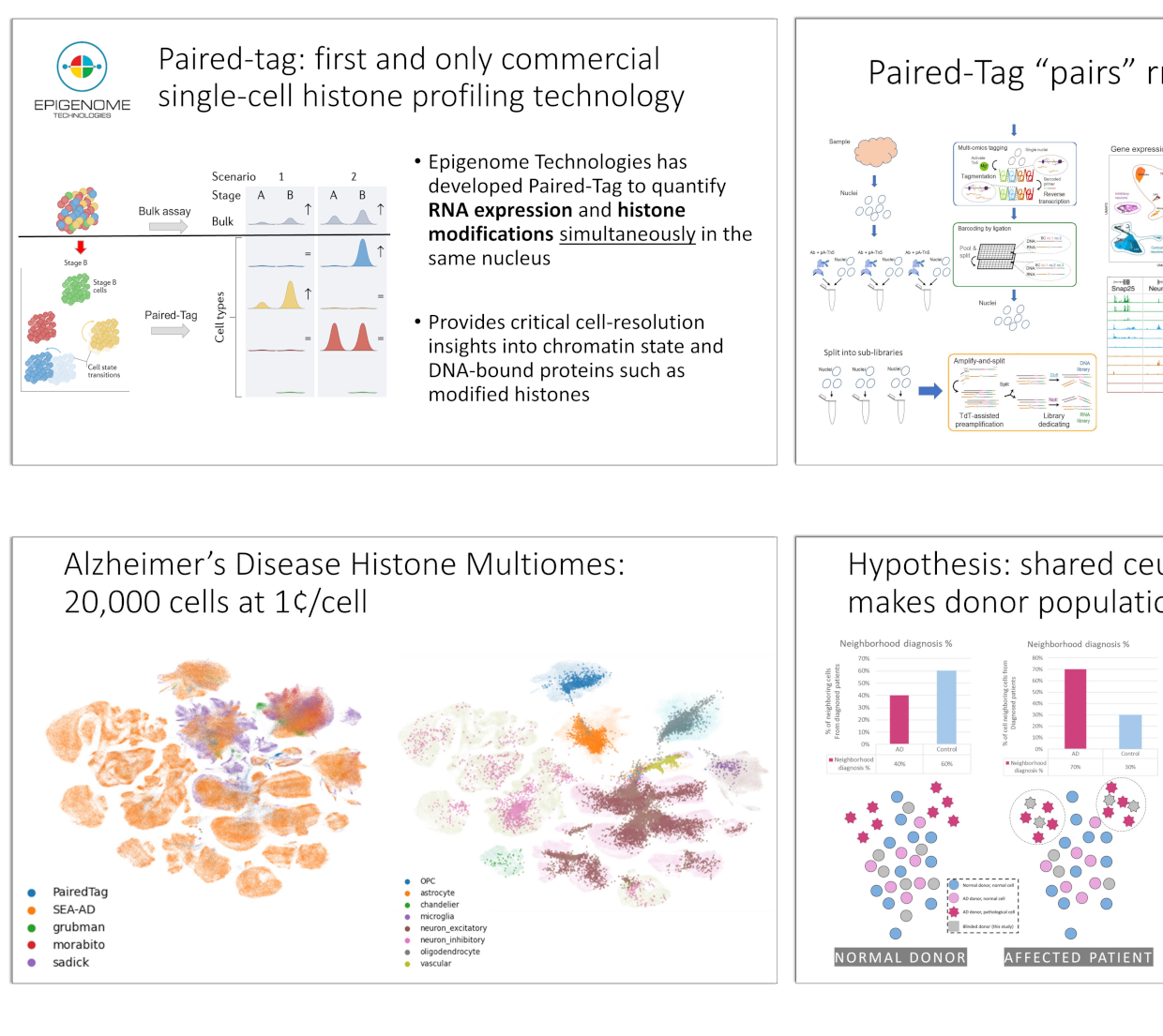
Paired-Tag: Single-Cell Epigenetic Profiling for Biomarker Discovery
Single-cell epigenetic profiling identifies key biomarkers of cellular dysfunction and disease.
Our methods simultaneously assess RNA expression and histone modifications in individual brain
cells and PBMCs, providing high-quality, reproducible data. We discovered novel cell-type-specific
cis-regulatory elements (cCREs) and heritability factors linked to Alzheimer's Disease (AD).
Additionally, our approach enables the identification of aging and disease-related biomarkers,
offering insights into their regulatory mechanisms.
Single-Cell Joint Analysis of Histone Modification State and Gene Expression
Applying Paired-Tag to Alzheimer's post-mortem brain tissue, we aligned with SEA-AD RNA data and identified shared pathologies. AD neurons showed H3K9me3 de-repression and H3K27ac activation of genes essential for neuronal identity and synaptic function, linking inflammation to dysfunction. Our single-cell epigenomic and transcriptomic analysis reveals specific cellular changes and potential therapeutic targets, demonstrating Paired-Tag’s effectiveness in understanding and treating complex diseases.
Download Poster (PDF)