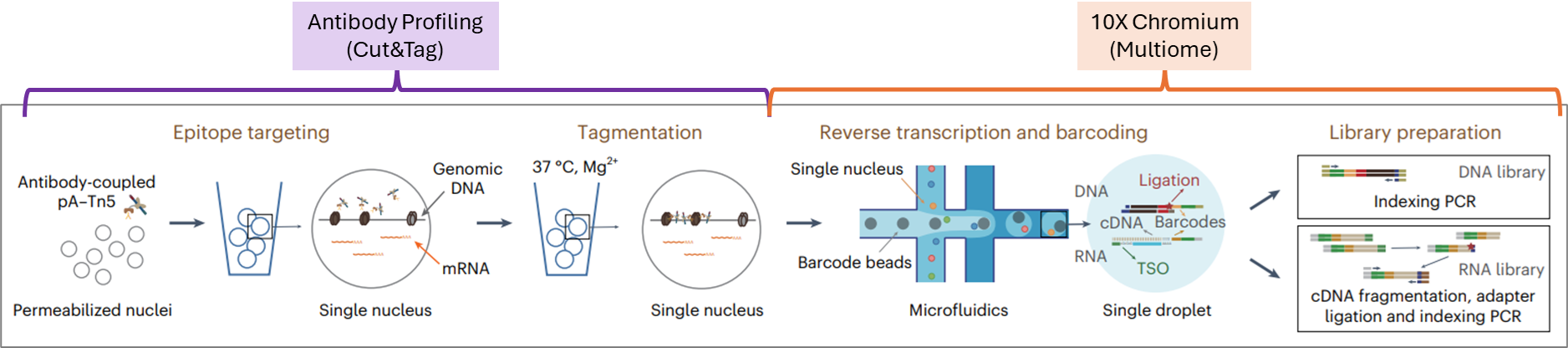
Paired-Tag Kits — Single-Cell CUT&Tag+RNA
Simultaneously measure RNA expression and chromatin modifications in the same cell. Profile histone marks, transcription factors, and gene expression together to reveal regulatory mechanisms without separate assays or flow sorting. Compatible with 10x Chromium workflows for seamless integration into existing single-cell pipelines.
Dual Readout
RNA + Protein
UMI Yield
5,000+ UMI/cell
Throughput Options
Standard + Multiplex
Choose Your Kit
| Product | Catalogue # | Platform | Size | List Price | |
|---|---|---|---|---|---|
| Droplet Paired-Tag | DP8101 | 10x Chromium | 8 Reactions | $2,470 | Order |
| Droplet Paired-Tag | DP16101 | 10x Chromium | 16 Reactions | $4,098 | Order |
| Multiplex Paired-Tag | MDP8101 | 10x Chromium | 8 Reactions | $2,580 | Order |
| Multiplex Paired-Tag | MDP16101 | 10x Chromium | 16 Reactions | $4,280 | Order |
Need guidance? Compare all kit options →
What You Need to Get Started
- Antibody: Validated primary antibody for target of interest (histone mark, TF, or chromatin protein)
- Single-Cell Platform: 10x Genomics Chromium system with ATAC or Multiome kit
- Reagents: 10x Genomics Multiome or ATAC reagents for barcoding and library prep
- Sequencing: Illumina-compatible sequencing system (NovaSeq, NextSeq, HiSeq)
Paired-Tag: Single-Cell CUT&Tag+RNA
Paired-Tag technology combines antibody-targeted chromatin profiling with RNA sequencing in individual cells, eliminating the need for separate scCUT&Tag and scRNA-seq experiments. By simultaneously capturing transcriptomic and epigenetic information, researchers gain direct insight into how gene regulation correlates with chromatin state across cell types and conditions.
Comprehensive Epigenetic Profiling
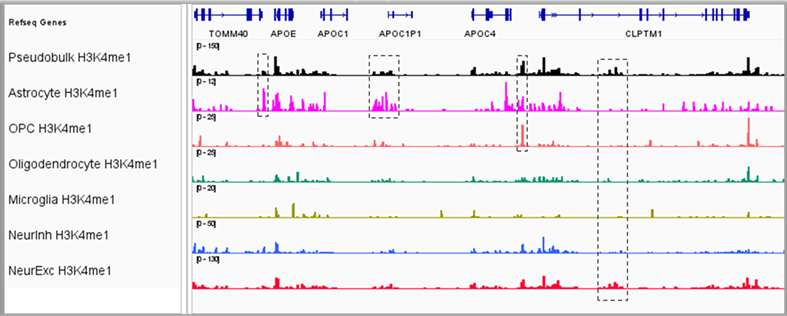
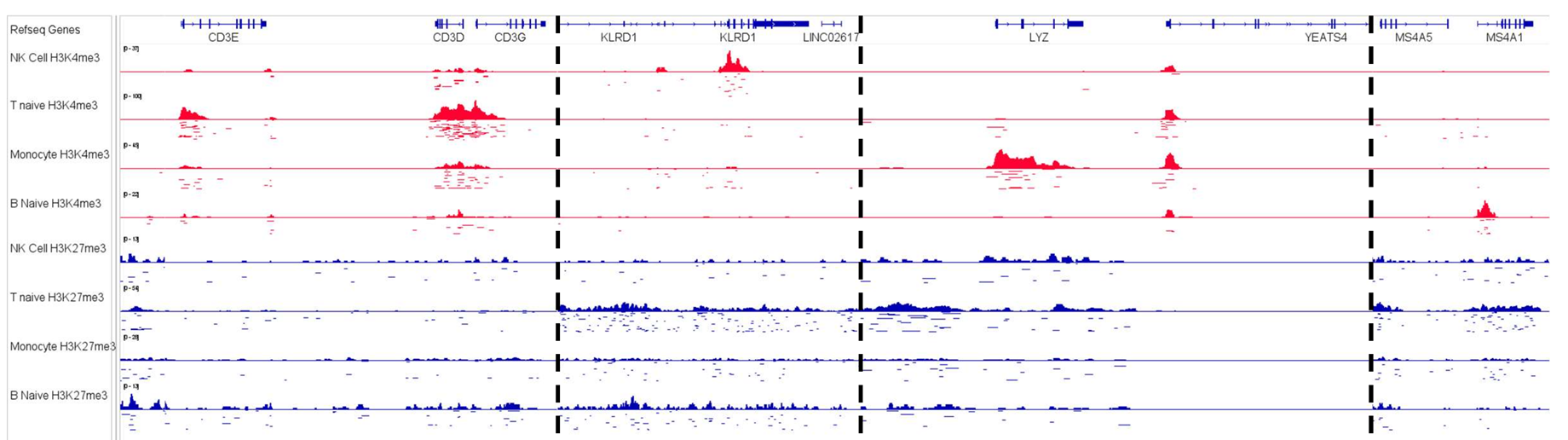
Target any validated antibody—histone modifications, transcription factors, or chromatin remodelers—while preserving RNA integrity. Profile H3K4me3, H3K27me3, H3K27ac, and other marks alongside gene expression to map regulatory landscapes. Goes far beyond ATAC-seq's open-chromatin view to reveal specific protein-DNA interactions driving cellular identity.
Seamless 10x Integration
Designed for compatibility with 10x Genomics Chromium workflows (ATAC or Multiome kits). Nuclei prepared with Paired-Tag reagents flow directly into standard 10x barcoding, eliminating custom hardware or workflow modifications. Analysis pipelines (Seurat, Signac, ArchR) process Paired-Tag data using established multiome methods.
Cell-Specific Insights Without Sorting
RNA-based annotation identifies rare populations, developmental stages, and disease states post-hoc—no flow cytometry required. Discover epigenetic signatures in cell types that lack surface markers or are too fragile for sorting. Ideal for tissues with heterogeneous cell composition where pre-enrichment would introduce bias or technical artifacts.
Research Applications
Researchers use Paired-Tag across developmental biology, cancer genomics, immunology, and perturbation studies to map how chromatin modifications and gene expression interact during cellular transitions, disease progression, and therapeutic response.
Developmental Biology
Track how histone marks and transcription factor binding evolve during lineage commitment. Map cell fate decisions by correlating chromatin accessibility changes with gene expression shifts across developmental time courses.
Cancer & Disease
Profile tumor heterogeneity by linking aberrant chromatin states to oncogene expression in single cells. Identify epigenetic drivers of therapy resistance and discover rare cell populations with stem-like chromatin signatures.
Immunology & Cell Fate
Characterize immune cell activation states through coordinated changes in regulatory marks and cytokine expression. Dissect how chromatin remodeling controls T cell exhaustion, macrophage polarization, and innate immune memory.
Choosing Between Droplet and Multiplex Paired-Tag
Both kits deliver simultaneous RNA and chromatin profiling, but differ in throughput strategy. Droplet Paired-Tag runs one assay per 10x lane for maximum cells per condition. Multiplex Paired-Tag uses barcoded enzymes to pool up to four assays in one lane, reducing per-sample sequencing costs.
| Feature | Droplet Paired-Tag | Multiplex Paired-Tag |
|---|---|---|
| Assays per 10x Lane | 1 assay | Up to 4 assays (pooled) |
| Cells per Reaction | 10,000 typical | 7,500 typical (per assay when multiplexed) |
| Total Throughput | 10,000 cells/lane | 30,000 cells/lane (4 assays × 7,500) |
| Cost per Assay | Standard sequencing cost | Reduced (shared sequencing) |
| Workflow Complexity | Standard | Requires sample pooling step |
| Best For | Deep profiling of single conditions; pilot studies | Multi-condition experiments; time courses; replicates |
Choose Droplet Paired-Tag If:
- Profiling a single experimental condition at maximum depth
- Running pilot experiments to validate antibodies
- Workflow simplicity is a priority
- Need maximum cells per sample (10,000+)
Choose Multiplex Paired-Tag If:
- Comparing multiple conditions, time points, or treatments
- Running replicate experiments in parallel
- Sequencing budget is constrained (shared lane costs)
- Comfortable with sample pooling and demultiplexing
Still Unsure?
Contact our scientists for a consultation. We'll help you select the optimal kit configuration based on your experimental design, budget, and throughput requirements.
Get Expert AdviceSafety Data Sheets (SDS)
Droplet Paired-Tag Kits
The following Safety Data Sheets apply to Droplet Paired-Tag products:
- Quench Buffer (CR1007)
- Incubation Buffer (CR1001)
- Activation Buffer (CR1006)
- Permeabilizer (CF1003)
- Stabilizer (CF1005)
- TAG Enzyme (CF1008)
- pA-Tn5 Adapter Complex (CF1001)
Multiplex Paired-Tag Kits (Additional)
Multiplex kits include these additional barcoded enzymes:
Request Protocols & Documentation
Access detailed protocols and technical documentation by registering with us:
Product Details
Droplet Paired-Tag — Standard Single-Cell CUT&Tag+RNA
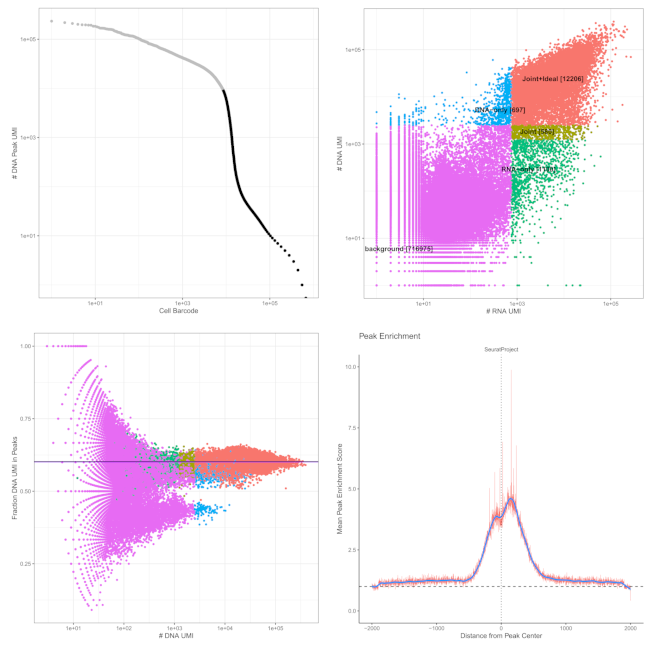
Droplet Paired-Tag captures chromatin modifications and RNA expression simultaneously in individual cells using antibody-targeted tagmentation. Nuclei are permeabilized, incubated with primary antibody and protein A-Tn5 fusion, then tagmented to insert sequencing adapters at antibody binding sites. RNA remains intact throughout, enabling dual profiling when processed through 10x Chromium ATAC or Multiome workflows.
Specifications
- Platform: 10x Genomics Chromium (ATAC or Multiome kits)
- Input: 100,000–500,000 nuclei per reaction (fresh or frozen)
- Typical Recovery: 10,000 barcoded cells per reaction
- Antibody Compatibility: Histone marks, transcription factors, chromatin remodelers
- Turnaround: 1 day for tagmentation, 1 day for 10x barcoding, standard sequencing timelines
- Sequencing Depth: 25,000–50,000 reads/cell (chromatin); standard scRNA-seq depth (RNA)
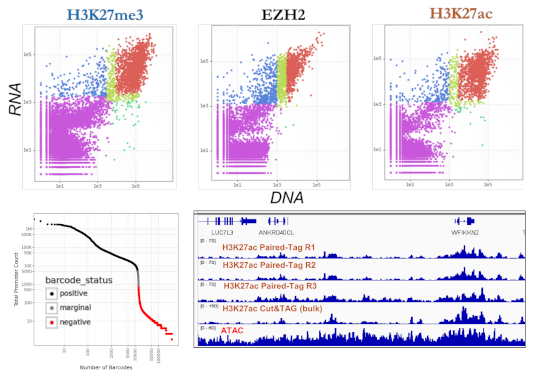
Performance Highlights
- Dual UMI Yield: Broad histone marks (H3K27me3, H3K4me1) deliver 5,000+ chromatin UMI/cell alongside standard RNA counts
- Tissue Versatility: Validated on brain, PBMC, organoids, tumors—fresh, frozen, or cryopreserved
- Reproducibility: Bulk and single-cell chromatin profiles show high correlation (R² > 0.9)
- RNA Integrity: Median gene detection matches standard scRNA-seq workflows
Key Advantages
- Simultaneous Profiling: Captures chromatin and transcriptome in the same cell, eliminating batch effects from separate assays
- No Flow Sorting Required: RNA-based cell type annotation post-hoc identifies rare populations without pre-enrichment
- Antibody Flexibility: Target any validated antibody—histone PTMs, TFs, or chromatin-associated proteins
- Standard Analysis Pipelines: Compatible with Seurat, Signac, ArchR, and other multiome tools
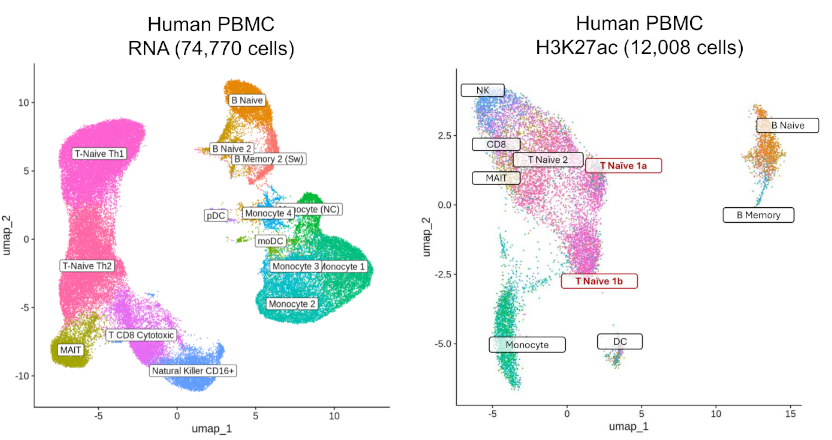
Biological Applications
- Cell Fate Decisions: Track how chromatin marks precede or follow gene expression changes during differentiation
- Rare Cell Discovery: Identify epigenetically distinct subpopulations within transcriptionally defined clusters
- Regulatory Mechanisms: Link specific histone modifications to target gene activation in cellular contexts
- Disease Heterogeneity: Profile aberrant chromatin states alongside oncogene expression in tumor samples
Best For
Single-condition experiments requiring maximum cell recovery per sample. Ideal for pilot studies validating antibodies, deep profiling of rare cell types, or projects where workflow simplicity is prioritized. Choose Droplet Paired-Tag when profiling one experimental condition at a time with 10,000+ cells per run.
Get Started with Droplet Paired-Tag
Available in 8-reaction and 16-reaction kits. Each reaction processes one antibody condition and captures approximately 10,000 cells when paired with 10x Chromium. Includes all tagmentation reagents, optimized protocols, and expert consultation.
Multiplex Paired-Tag — High-Throughput CUT&Tag+RNA
Multiplex Paired-Tag extends the standard workflow with uniquely barcoded Tn5 enzymes, enabling up to four independent assays to be pooled in a single 10x Chromium lane. Each antibody condition receives a distinct enzyme barcode during tagmentation. After pooling, samples are processed together through 10x barcoding and sequencing, then computationally demultiplexed. This approach dramatically increases throughput while reducing per-sample costs.
Specifications
- Platform: 10x Genomics Chromium (ATAC or Multiome kits)
- Multiplexing Capacity: Pool up to 4 assays per 10x lane
- Input per Assay: 75,000–150,000 nuclei per reaction
- Typical Recovery: 7,500 cells per assay (30,000 total when 4 assays pooled)
- Enzyme Barcodes: Individual molecules enzymatically tagged, enabling full use of doublet tagmentation.
- Demultiplexing: Computational separation based on enzyme barcode sequences

Performance Highlights
- Preserved Data Quality: Barcoded enzymes enable accurate demultiplexing with minimal crosstalk (<2%)
- Cost Efficiency: Shared sequencing lane reduces per-assay costs by ~75% vs. separate runs
- Throughput Scaling: 30,000 total cells when pooling 4 assays (7,500 each) in one lane
- Balanced Recovery: Cell yield consistent across all four barcoded conditions
Key Advantages
- Multi-Condition Profiling: Compare time points, treatments, replicates, or different antibodies in parallel
- Reduced Reagent Costs: Share 10x barcoding reagents and sequencing across four samples
- Faster Turnaround: Process multiple conditions simultaneously rather than sequentially
- Flexible Experimental Design: Pool 2, 3, or 4 assays depending on cell yield and sequencing depth requirements
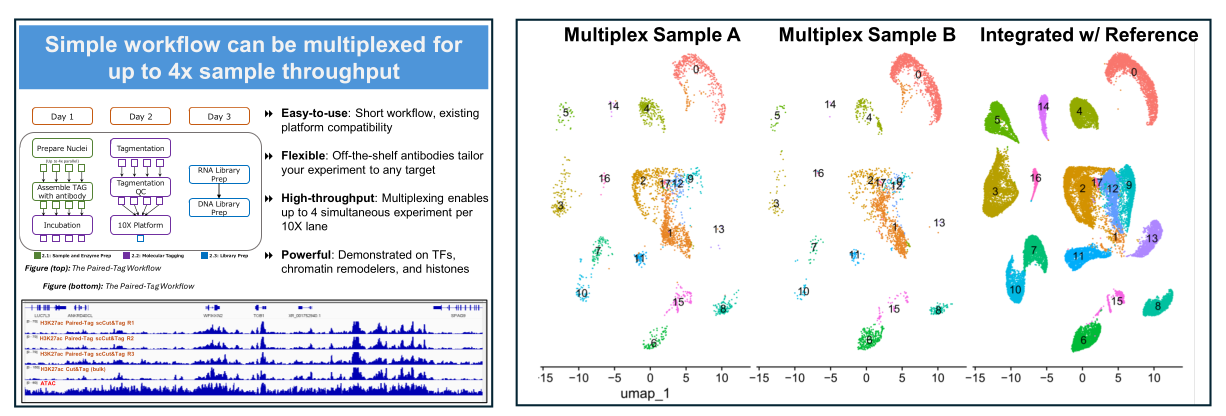
Biological Applications
- Time-Course Studies: Profile chromatin and RNA changes across developmental or treatment time points
- Multi-Condition Comparisons: Compare disease vs. control, treated vs. untreated, or multiple cell types
- Antibody Validation: Test multiple antibodies simultaneously to identify optimal targets
- Replicate Experiments: Reduce batch effects by running biological replicates in the same lane
Best For
Multi-condition experiments where comparing groups is critical—time courses, dose responses, case-control studies, or replicate profiling. Ideal when sequencing budget is constrained or when reducing batch effects between conditions is paramount. Choose Multiplex when you need to profile 2–4 related samples and can tolerate slightly lower cells per condition (7,500 vs. 10,000).
Get Started with Multiplex Paired-Tag
Available in 8-reaction and 16-reaction kits. Each kit includes four barcoded Tn5 enzyme variants, enabling multiplexed experiments. Comprehensive protocols cover pooling strategies, QC checkpoints, and demultiplexing workflows.
Ready to Profile Chromatin and RNA Together?
Start with a kit for immediate hands-on experiments, or let our experts handle the complete workflow as a managed service.