Simultaneous RNA and Chromatin Profiling at Single-Cell Resolution
Epigenome Technologies delivers Paired-Tag programs that profile histone modifications, transcription factors, and gene transcription from the same cell without computational integration. We qualify antibodies, optimize nuclei handling, and return dual cell-by-peak and cell-by-gene matrices that reveal how chromatin state drives transcriptional heterogeneity.
True Multi-Modal Measurement
Chromatin and RNA from the same cell
Built for Cellular Heterogeneity
Cell-state resolution preserved
Scalable Study Design
From pilot to cohort
Paired-Tag: Dual-Modality Single-Cell Profiling
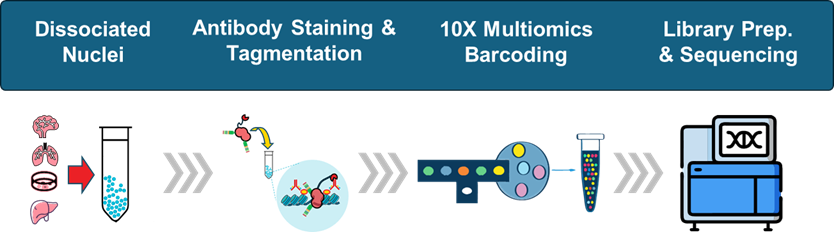
Paired-Tag simultaneously captures histone modifications, transcription factors, or chromatin remodelers alongside full-length transcript sequences from individual cells. The dual readout reveals how chromatin state drives gene expression decisions without requiring computational integration or multi-sample inference.
Where teams use the program
- Mechanistic cancer biology linking chromatin state to tumor heterogeneity and treatment resistance.
- Immunology studies characterizing how epigenetic remodeling enables cell-state transitions.
- Developmental biology tracking chromatin-to-transcript coordination during differentiation.
- Rare-cell epigenetics without FACS sorting or enrichment biases.
- Perturbation validation studies measuring on-target chromatin and off-target transcriptional effects.
Collaboration model
- Joint design sessions align target antibodies, cell counts, sequencing depth, and platform selection.
- Antibody validation runs confirm signal-to-noise before full cohort commitment.
- Project scientists remain embedded through QC reviews, data interpretation, and reporting.
Included interpretation
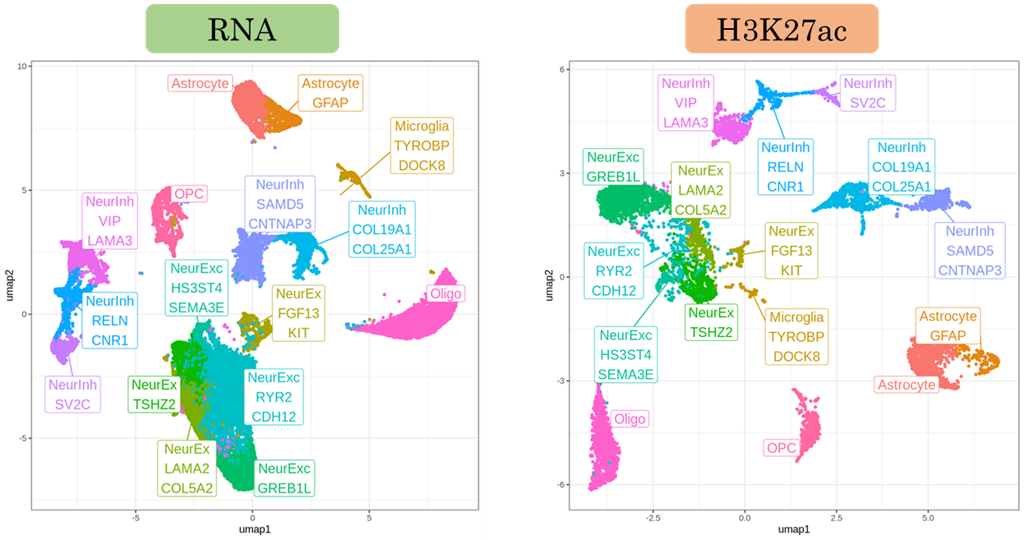
- Cell-type clustering and dual epigenetic-transcriptional signature annotation.
- Chromatin-to-gene linkage analysis revealing direct regulatory relationships.
- Differential epigenetic and expression analysis with joint visualization.
- Actionable slide-ready visualizations delivered alongside raw outputs.
Aligning platform to throughput and study design
| Platform | Cat. No. | Best For | Throughput | |
|---|---|---|---|---|
| Droplet Paired-Tag | DPS201 | Focused discovery; single-sample or small cohorts with deep characterization | 1 sample per 10x lane; 5,000–10,000 cells/sample | Get Quote |
| Multiplex Paired-Tag | MDPS201 | Large cohorts and high sample throughput; patient/donor multiplexing via barcoding | 8–12 samples per run; 500–2,000 cells/sample | Get Quote |
Droplet Paired-Tag (Standard Throughput)
- Dedicated 10x lane per sample for maximum signal-to-noise
- Recommended for small and/or low-input cohorts.
- Highest per-cell sequencing depth and fragment complexity
Multiplex Paired-Tag (High Throughput)
- Custom oligo barcoding for unambiguous sample demultiplexing
- Ideal for cohorts with 4+ samples or translational studies
- Up to 8K nuclei / sample; 30K nuclei / lane.
Transparent workflow and checkpoints
-
Plan & validate antibodies
We confirm target antibodies, cell counts, barcoding strategy (if multiplexed), and nuclei handling; optional antibody validation runs ensure signal quality before full cohort commitment.
-
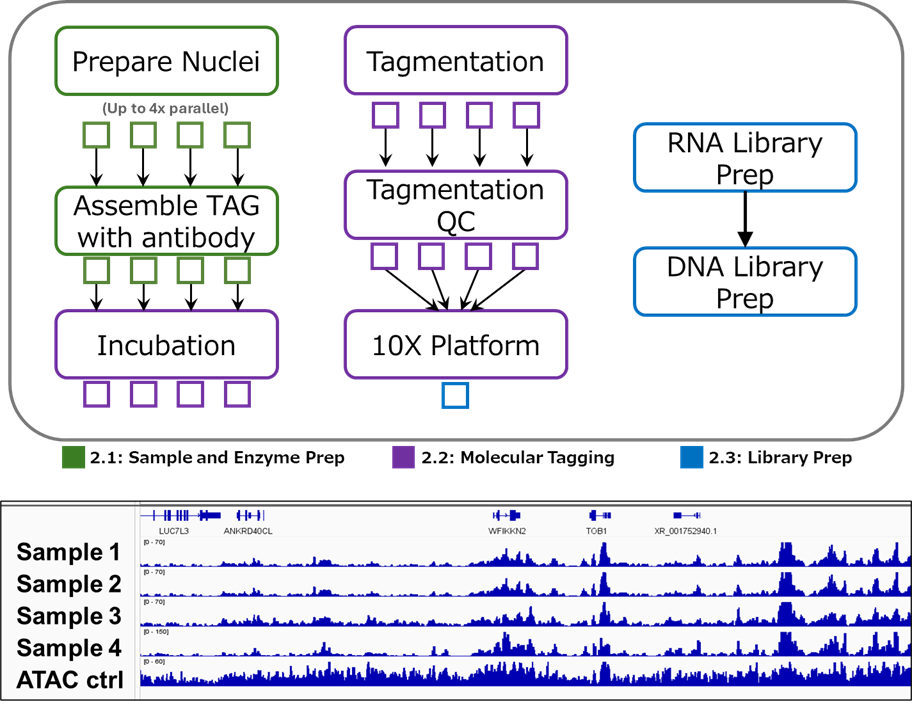
Prepare & construct
Nuclei isolation, antibody staining, RNA capture, and droplet encapsulation with QC on viability, tagmentation efficiency, RNA complexity, and barcode distribution.
-
Sequence
Paired-end runs on NovaSeq or NextSeq platforms; multiplexed samples demultiplexed via cell-hash barcodes with per-sample lane balancing.
-
Interpret
Automated cell calling, peak annotation, transcript quantification, and chromatin-transcript linkage analysis with data packages aligned to your preferred environment.
Operational cadence
- Day 3 antibody validation QC review (if requested) with your project scientist.
- Day 8 nuclei prep and library construction status with preliminary fragment and RNA complexity profiles.
- Day 14 library acceptance with tagmentation efficiency, RNA yield, and barcode distribution reports.
- Day 18 FASTQ, cell-by-peak, and cell-by-gene matrices delivered; day 20 interpretive briefing.
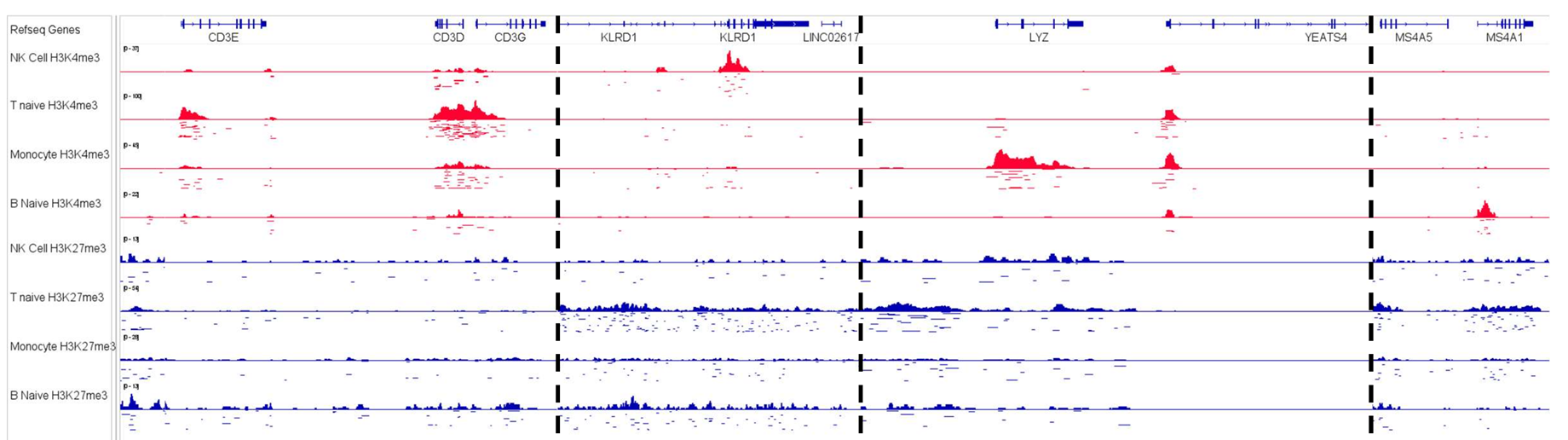
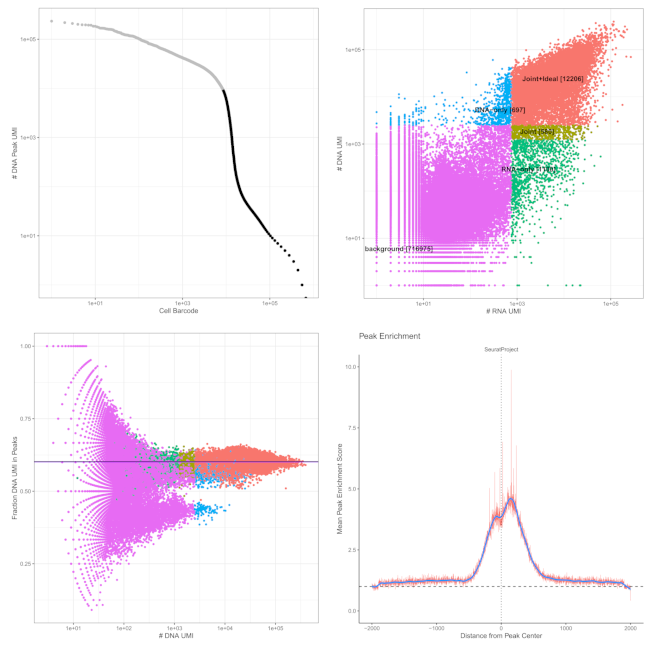
Deep epigenetic insight
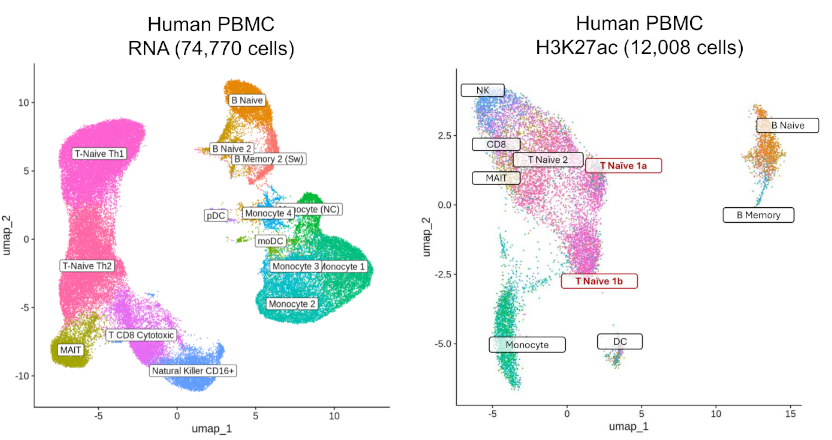
High Quality Data
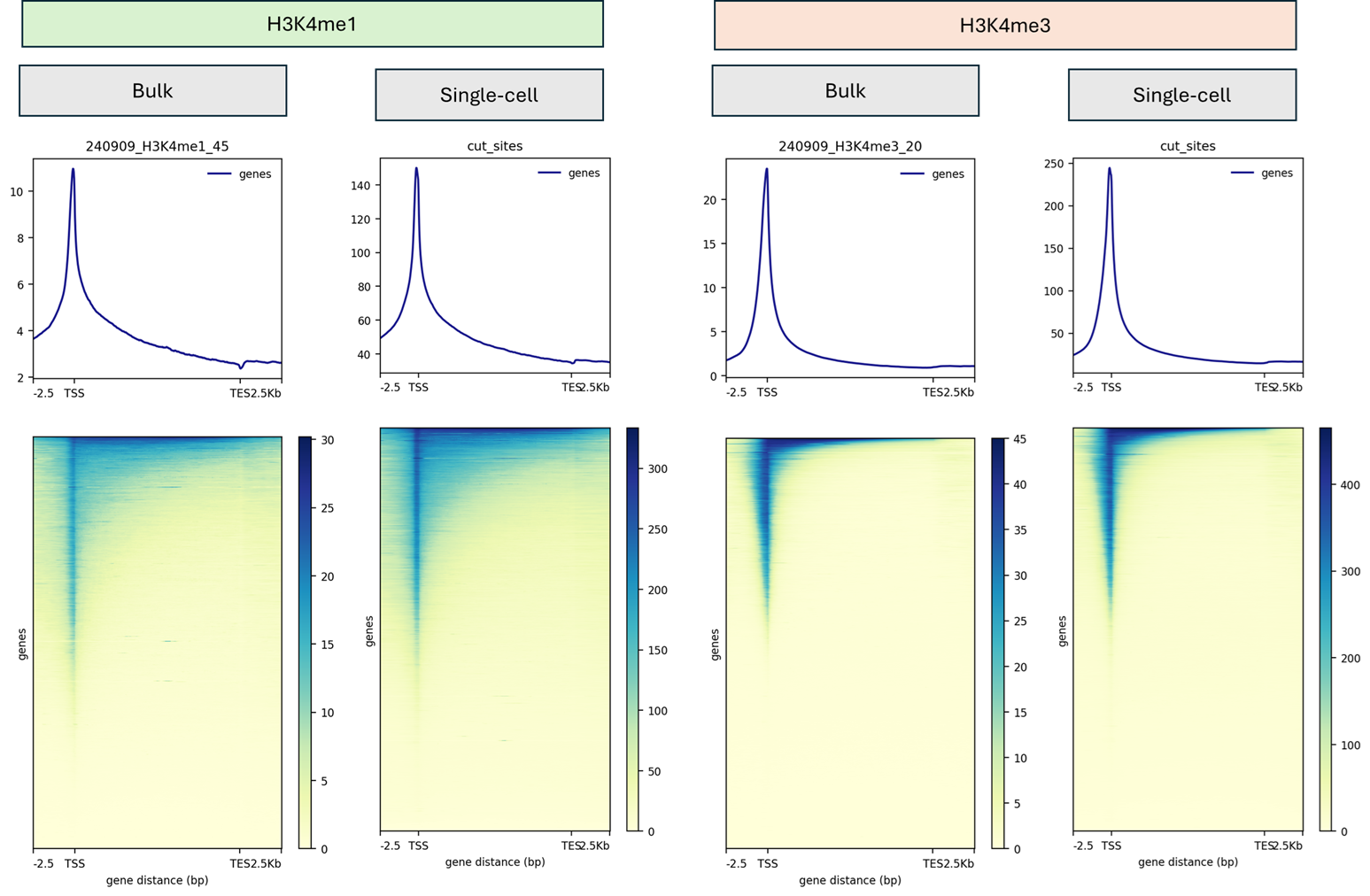
- TSS enrichment distributions with cohort medians and antibody-specific benchmarks.
- RNA mapping rates and gene detection summaries reviewed prior to sequencing approval.
- Chromatin fragment size and nucleosome phasing profiles for every sample.
- Cell-barcode complexity and background noise assessments with per-sample validation.
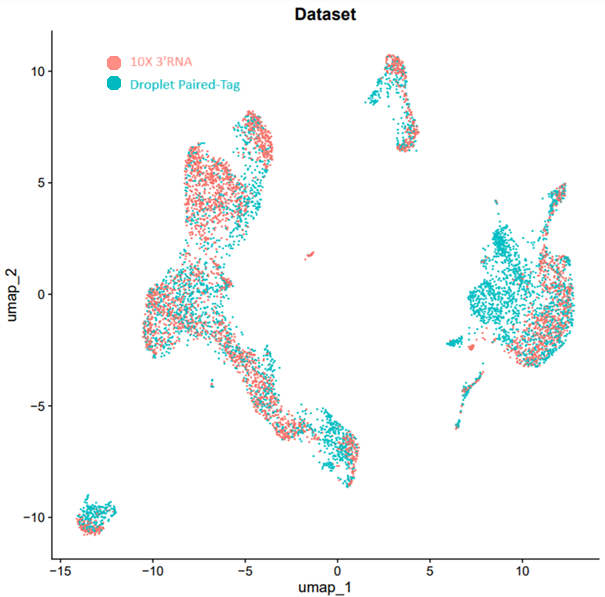
- Dual-modality concordance metrics showing chromatin-transcript linkage quality.
Delivery package
- Sequencing-ready libraries with concentration, fragment distribution, and complexity documentation.
- FASTQ files, cell-by-peak matrices, cell-by-gene matrices, and metadata formatted for Seurat, Scanpy, or custom pipelines.
- Interpretive report summarizing cell-type clustering, dual differential analysis, chromatin-transcript linkage, and regulatory annotations.
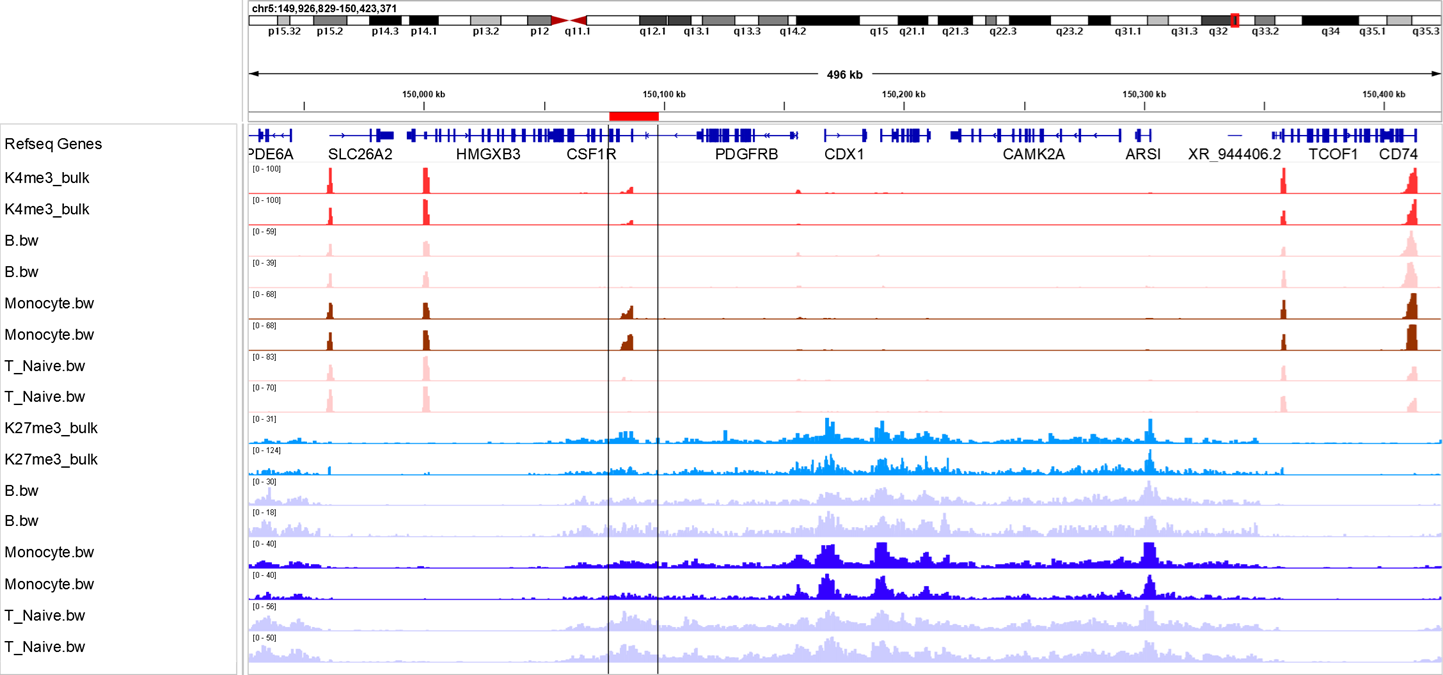
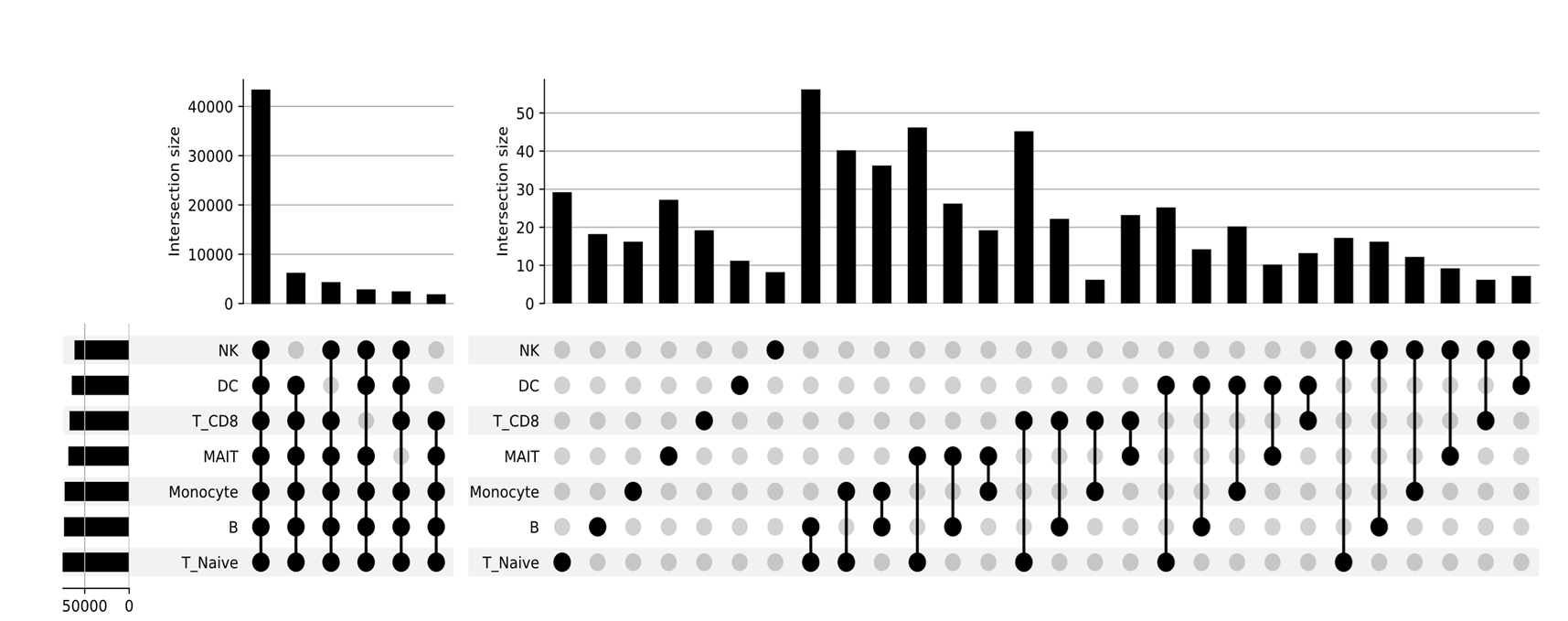
Pancreatic Islet Cell Heterogeneity in Diabetes
Paired-Tag profiling of pancreatic islets has revealed novel insights into β-cell heterogeneity and stress responses directly relevant to diabetes pathogenesis. The assay uncovered distinct β-cell subtypes derived from biochemically and epigenetically defined progenitors, demonstrating how histone modification patterns and epigenetic dosage shape functional specialization in insulin-secreting cells.
Key Findings
- Identified β-cell subtypes with distinct epigenetic and transcriptional signatures
- Mapped epigenetic dysregulation underlying β-cell stress during autoimmune attack in Type 1 diabetes
- Revealed regulatory networks far more detailed than bulk approaches
- Advanced target identification for therapeutic intervention
Prenatal E-Cigarette Exposure and Brain Development
A recent study applying Paired-Tag to the prenatal mouse brain exposed to e-cigarette aerosols delineated the epigenetic consequences of environmental exposure on neurodevelopment. The assay revealed altered histone modification landscapes at enhancer and promoter regions within excitatory neurons and glia, associated with disrupted gene expression programs critical for brain development.
Key Findings
- Mapped cell-type-specific epigenetic vulnerabilities to prenatal exposures
- Linked chromatin changes directly to transcriptomic dysregulation
- Identified molecular pathways mediating environmental toxin impact
- Revealed mechanistic basis for potential long-term neurodevelopmental impairments
Partner with our scientists
Share your target antibodies, cohort size, and required timelines. We will return a scoped Paired-Tag brief outlining platform selection (Droplet vs Multiplex), antibody validation strategy, multiplexing approach (if applicable), QC checkpoints, and downstream reporting.
- Sample requirements: 100K–200K nuclei preferred; low-input contingencies available upon consultation.
- Storage guidance: Fresh or cryopreserved nuclei accepted with documented handling.
- Data options: Raw FASTQ, cell-by-peak/gene matrices, and interpretive outputs available individually or bundled.
- Support: Project scientists provide experimental planning and guidance, data reviews, and troubleshooting.